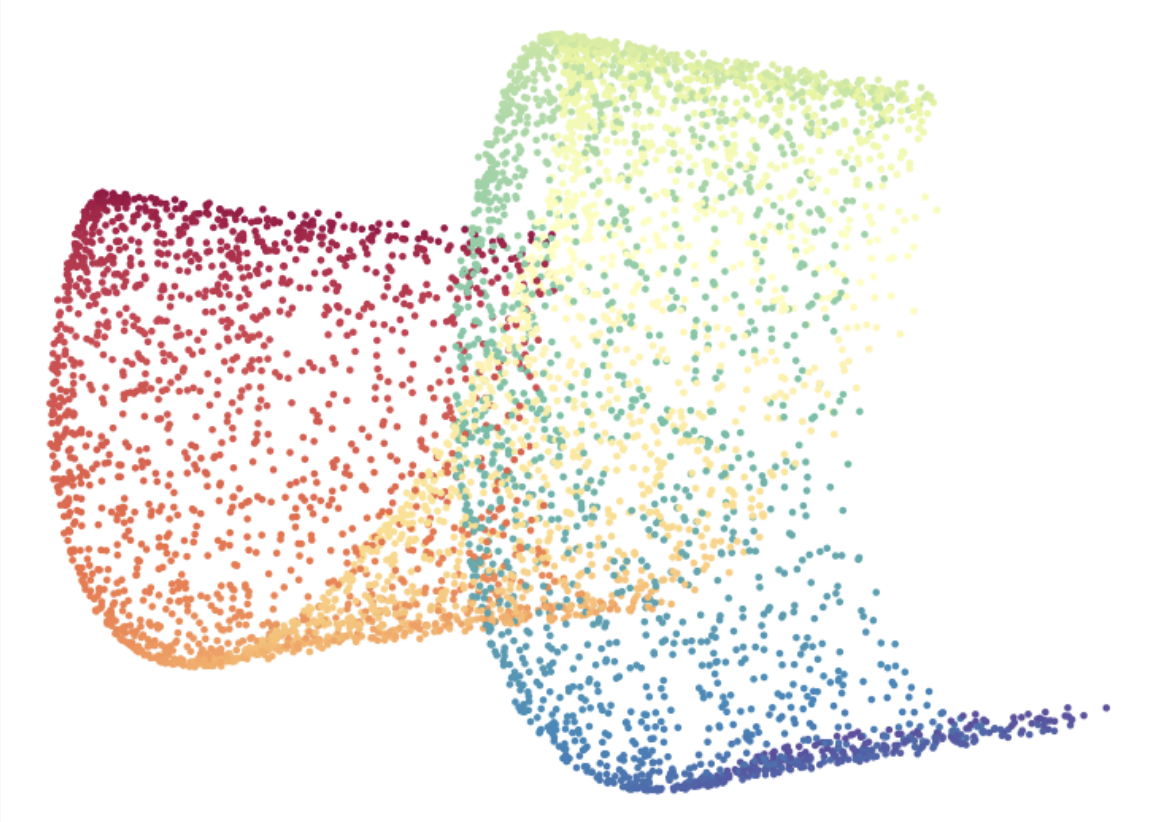
Linearized Diffusion Map
We introduce the Linearized Diffusion Map (LDM), a novel linear dimensionality reduction method constructed via a linear approximation of the diffusion-map kernel. LDM integrates the geometric intuition of diffusion-based nonlinear methods with the computational simplicity, efficiency, and interpretability inherent in linear embeddings such as PCA and classical MDS. Through comprehensive experiments on synthetic datasets (Swiss roll and hyperspheres) and real-world benchmarks (MNIST and COIL-20), we illustrate that LDM captures distinct geometric features of datasets compared to PCA, offering complementary advantages. Specifically, LDM embeddings outperform PCA in datasets exhibiting explicit manifold structures, particularly in high-dimensional regimes, whereas PCA remains preferable in scenarios dominated by variance or noise. Furthermore, the complete positivity of LDM’s kernel matrix allows direct applicability of Non-negative Matrix Factorization (NMF), suggesting opportunities for interpretable latent-structure discovery. Our analysis positions LDM as a valuable new linear dimensionality reduction technique with promising theoretical and practical extensions.