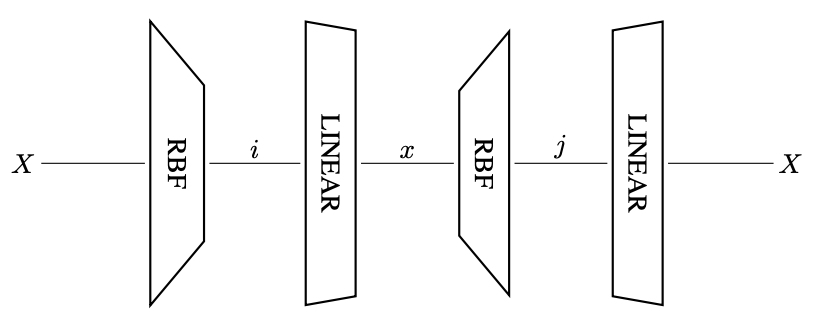
Linearized Diffusion Map
We introduce the Linearized Diffusion Map (LDM), a novel linear dimensionality reduction method constructed via a linear approximation of the diffusion-map kernel. LDM integrates the geometric intuition of diffusion-based nonlinear methods with the computational simplicity, efficiency, and interpretability inherent in linear embeddings such as PCA and classical MDS.